|
|
BIO 432
Field Botany
Phil
Ganter
301 Harned Hall
963-5782 |
| Restinga Jurubatiba
in Brazil The restinga
biome (a tropical dune forest) is both valuable (it contains a diversity
of valuable plants and animals) and disappearing. The restinga is part
of the Atlantic Coastal Forest biome in Brazil, one of the most diverse
terrestrial plant communities. Less than 10% of the original area occupied
by this biome is undisturbed by man
|
Systematic Botany and Taxonomy
Email
me
Back
to:
Why this
note?
In this course, we will
be identifying plants. This means more than linking a specimen to a common
name or to a Latin binomial (the genus - species tag often called the "scientific
name"). We will use the system developed by biologists to organize organisms
into a logical scheme. As we will see, there are two schemes common in science.
Below is the organization used for this essay done as links to the various
sections of this page. The purpose of the page is to both introduce you to
classification of living organisms and to avoid a practical problem that
arises
from the diverse methods in use to classify.
This page deals with classification.
Biological classification is the process of grouping and naming the many different
sorts of living organisms. It is not an easy or straight-forward task and, although
biologists have done it since the origins of biology (and, earlier, the origins
of natural history -- Aristotle was interested in biological classification),
the task is never ending. There are both new organisms being discovered constantly
and there are new ways of classifying being devised. This page introduces two
ways of classifying organisms: Taxonomy and Systematics.
Taxonomy
is the science of naming organisms based on accepted criteria (the criteria
are derived from the scheme underlying the taxonomy). This sounds pretty
cut
and dry but, as in many things, the devil is in the details. As a biologist,
you should be interested in the science of taxonomy, even if you have
never
given it serious consideration or have done so and dismissed it as uninteresting
(trivial? prehistoric? fussy?). You certainly make use of taxonomy. We
cannot
make a systematic study of things (living or otherwise) without a system for
organizing our knowledge. There are just too many things. Taxonomy happens
whenever
the number of things gets too large to allow them to be discussed clearly.
It happened to geneticists. A consistent naming scheme for loci and alleles
is often
adopted by those studying a particular model organism after the number of genes
identified has grown large. It even happened to atomic physicists
after the atom
smashers
started
to
find
heaps of
subatomic
particles. (By the way, Enrico Fermi, an eminent physicist, remarking on the
number of newly described atomic particles, supposedly remarked that,
if he could remember the names of the particles, he would have become
a botanist). Certainly, any biologist working in natural systems is in
great need
of some taxonomy.
Anyone
can see
that
there are lots
of different plants, even in simple communities. The question considered
here
is "how should we go about organizing groups of living things?"
One way of organizing knowledge about differences
between plants is to name the plants in such a way that their names convey some
additional information beyond the name. Certainly, taxonomists are not satisfied
to merely label organisms. This point may not be so obvious. Suppose you have
a friend who has just acquired two dogs that he named Bob and Larry. One is
large and aggressive and the other small and gentle. When you see the two dogs,
could you tell which is Bob and which is Larry? No. In this case, the names
convey no information and merely serve as tags to identify the dogs. Suppose
your friend had named the dogs Killer and Daisy. When you saw the same two dogs
with these names, you could make an informed guess as to which was Killer and
which Daisy (assuming you know your friend well enough to know if he or she
is a wise guy or not). So, names can be more than labels. They can convey additional
information. Taxonomy gives names to organisms that convey information about
the organism. If I tell you that the cactus in the picture below is Pilosocereus
royenii, I have given you a wealth of information about the organism in
the picture.

I have told you something
about its external features (all Pilosocereus have similar flowers,
similar overall growth form, and similar tufts of hairs along the ribs
where they will produce flowers), its
internal features (all Pilosocereus have similar secondary chemistry
and anatomic details), and where it comes from (P. royenii is only
found on some Caribbean islands). Of course, you have to know something about
the
details of naming
cacti to know all of this, but once known, names almost become stories about
organisms, not just names. Recently, a friend sent me a picture of someone
that
had cacti in the background. From the cacti, I could tell him where the picture
was taken (the tip of the Baja California peninsula). Taxonomy is constantly
used in diverse fields, such as forensic medicine, agriculture, civil engineering,
and landscaping.
Taxonomy
and evolutionary biology have
become linked, although each has an independent origin and history.
As evolution became accepted
by the the majority of biologists, taxonomy and evolution became fused into
the field of systematics. Systematics
is the science of naming and organizing organisms based on common ancestry
(= evolutionary relatedness). In a systematic taxonomy, all of the species
in a
genus are more related (= have a more recent ancestor in common) to one another
than to any other species. Most taxonomies used today are systematic taxonomies,
although some persist that are not. (If you think of the reptiles as a taxonomic
group, then non-systematic taxonomy persists in you!). An example of
scientists
using non-systematic taxonomy comes from the old standards for
describing fungi (this has been changed in the new Botanical Code). When
describing a new yeast, if I could
not find any sexual characteristics, then I had to place the species into a
genus with other asexual species (the familiar yeast genus Candida is
an example of an asexual genus), even if gene sequence data indicate that
the
new
species is very closely related to a sexual species and not closely related
to any other member of Candida. The asexual genus was a hodgepodge
of unrelated species. Thus, much of fungus taxonomy
was not
systematic.
When describing an sexual yeast, a systematic approach
was followed but not when the new yeast was asexual. This
inconsistency was the product of history. Until genetic data became widely
available, it
was
not clear to which sexual genus many asexual species belonged so it was a practical
decision to just put them into a "hodgepodge" of a genus. In
2012, new rules based on genetic methods were instituted to allow a systematic
approach to all fungi.
Often, systematic taxonomies are referred to as "natural
classifications." This means that the taxonomy
reflects groups that exist in nature. The taxonomist is just
describing groups that already occur. If you have a group of
cats to classify, placing all lions in one group,
all house cats in a group, and all tigers in another is a natural classification.
However, perhaps you are classifying the cats at a zoo and
you are in charge
of feeding them. In this instance, becuse the kind of food given might
differ based on the size of the cat, placing all cats under 30 pounds in one
group and all cats over 30 pounds in another
is
a useful
taxonomy
but it is
an artificial grouping (ocelots and lynx in one group, tigers and lions in
another), not a natural grouping. Non-natural
classifications are the result of decisions made by the taxonomist but they
do not reflect
the
natural
situation. On
way to think of the difference is reflected in the statement "we discover
natural groups but we invent artificial ones."
Usually, you see a scientific name reported as a binomial
(which simply means a two-part name), like Syringia vulgaris (the
garden lilac). Latin and Greek are used (and a good binomial doesn't
mix the two). When a term is not Latin or Greek it is Latinized. (Opunita
engelmannii
was named for a botanist named Engelmann). This makes the scientific
name a foreign term in English and so scientific names are italicized (as are
all
foreign
terms in English). However, the binomial is really only the final two
parts of a series of names, as the first part is the name of the genus to which
the
organism belongs and the second is its species. Perhaps we use the binomial
because we often use two names for people and using two for an organism then
seems to identify the organism as though it were a person. This is a
bit misleading. When we name a person John Doe, we are attempting to
give an individual a
name
that belongs to no one else. When we call an oak tree Quercus emoryi,
we mean it belongs to the genus and species named. There are many Quercus
emoryi trees and a different scheme is needed to identify individual
plants.
Traditionally, taxonomists developed a hierarchical
scheme for naming organisms. A hierarchy is an organizational scheme based
on a series of levels. Each level has one or more groups (by definition,
the highest
level has only one group). At any level, each group is composed of groups from
the next level down. Thus, a genus like Quercus (the oaks)
is composed of groups from the next level down (species in the case
of a genus). All of this might
seem so obvious as to be trivial. However, hierarchies are not the only
way to organize things (although they seem to be the scheme of choice for many
areas
of human knowledge). We will not spend time here on other ways to organize
but biologists have made use of them. Behavioral biologists have discovered
that
some animal societies are strictly hierarchical and that others have little
hierarchy, although they do not lack organization. Ecologists have proposed
that some communities of competing species might be more stable if they are
not organized hierarchically.
There is more to traditional taxonomy than its hierarchical scheme.
How groups are formed is key to understanding the plan and understanding
the
plan is key to getting the information available from a scientific name. Taxonomic
groups are formed on the basis of members being similar to one another. This
similarity is not a vaguely defined thing but is based on particular characteristics
which all members of a group must share. Such characteristics are called key
characteristics. Thus there are specific
key characteristics that all Quercus emoryi have and generic key
characteristics all Quercus have. A northern red oak (Quercus
rubra) shares
generic key characteristics with an Emory oak (Quercus
emoryi) but not specific key characteristics (note that
I use the adjective "specific" to mean "having to do with
biological species" and not the more usual "having to do with
this particular thing"). Ideally, key characteristics should
be unique to that group. This is sometimes difficult to do but in any
case, key characteristics
must belong
only to the members of a one group (say one species) within a larger group
(the genus to which that species belongs).
Descriptions of species in
books and on the web often have many characteristics listed that are not
key
characteristics. My tree identification book says northern red oaks have
leaves 5 to 8 inches long. It also says swamp chestnut oaks have leaves
with the same
range of lengths, so leaf length is not a key characteristic for either species
of oak. Key characteristics can be found along with a host of other
characteristics
of the group in the scientific article that first defined the group or the
most recent article that re-defined the group. (When we learn more about
the organisms,
redefinition is often necessary). They are often difficult to use unless
you are in a laboratory as they can be microanatomic or chemical characteristics.
When identifying a plant, you will usually use a series of non-key characteristics
to decide on membership in a species. So, if you have four different
oak species,
one may have 5 inch leaves and smooth acorn cups, another may have 5 inch leaves
and rough cups, a third may have 6 inch leaves and rough cups and a fourth
6
inch leaves and smooth cups. Neither leaf size or acorn cup texture is
a key characteristic because no group has a unique leaf size or cup texture. Even
so, these characteristics can be used in combination to separate the four species.
| |
Leaf
Size |
| 5
inch |
6
inch |
| Cup
Type |
rough
cup |
Species
A |
Species
B |
| smooth
cup |
Species
C |
Species
D |
In addition to defining key characteristics, taxonomists try to maintain
consistency when describing new
taxa, whether they are families, species or genera. By consistency,
I mean that they prefer to organize species into genera or individuals into
species such
that the level of similarity between species within each genus or individuals
within each species is the same. Imagine you are describing eight different
species that do not belong to any known genus. But how many new genera
to form? You feel that the most unlike of the eight species are too dissimilar
to
belong
to the same genus so more than one new genus will be necessary. Upon
inspection, you find that there are two clusters of new species (one with three
species and
the other with four) that are distinct from one another but with little difference
between species within each cluster. These seven species will form
two new genera
but what to do with the eighth, which is not very similar to the other seven
species? Usually, it will become a "monospecific" genus rather
than just being included in with the genus to which it is most similar.
Thus, the new genera described have consistent differences between
the species. Consistency can be hard to maintain. Suppose you
discover an ninth species in this group. It is most
similar to the eighth species that is now in its own genus, but the level
of similarity
between the new species and the eighth is not as high as the level of similarity
within the other two genera. If you include the new species in the
same genus
with the eighth, consistency has been undermined because now one genus has
somewhat more dissimilar species in it than the other two genera. If
you describe a fourth genus,
the number of genera, etc. will seem to grow very quickly. The number
of names generated is a concern because we simply can't keep track of an infinite
number of names, so generating to many new names defeats the an important goal
of taxonomy.
Consistency is very hard
to maintain when the number of species in a group gets large or when describing
different types of organisms. The morphological differences between the
species of the genus Homo (to which you belong) are large when compared
to the differences between species within frog genera. Consider the
difference in size
and shape for the skulls of H. neanderthalensis versus H. sapiens. Frog
species with such large morphological differences as there are between species
of Homo
are usually put into different genera or even different families. The
rules for separating frogs into different species are different from those
for separating
primates into species, which is not consistent. A second problem for
the goal of consistency is that different kinds of characteristics may support
the
formation
of different taxa. The morphological difference between man and chimpanzee
is so great that they are not part of the same genus but the genetic distance
(measured as the average difference between the
basepair sequence for a sample of genes from each species) is small. Most
genes that vary at all do so only by a few percent of the basepairs. Many
vertebrate species include individuals that are genetically more dissimilar
than the
average
man and chimp. The growing use of gene sequence data often supports previously
defined taxa but there are important cases in which the new data is not consistent
with older data. What to do then?
The problem goes even deeper
than what to do when different types of data disagree. Notice that
in the explanation of what taxonomists do above, there is a lot of decision
making by the taxonomists.
The researcher must decide if all characteristics are equally important
or if some characteristics carry more relevant information. When examined
in detail,
it becomes obvious that decisions must be made at each step of the process
of constructing a taxonomic scheme. This has long bothered many who
study organisms
above the level of the individual. Given the same set of data, two taxonomists
might easily decide to group the organisms differently. Different
conclusions
from the same data is problematic. There have been two general responses
to this problem: phenetics and cladistics.
The first, phenetics, is not a systematic approach (that it, based on
relatedness), but the
second one, cladistics, is systematic.
Pheneticists approach taxonomy by measuring as many characteristics
of each individual as possible. They collect large data sets in which
there are
bound to be inconsistencies. They then apply a mathematical algorithm
designed to form groups from this large mass of data. The math models
have become both
powerful and complex as the field has developed. They have the useful
property that, given the same data, two different pheneticists will form the
same groups
if both use the same algorithm to do the grouping. Consistency is still
not achieved because there are lots of models, which means that the pheneticist
must make a decision about which model to use and this is a bit more decision
making than purists will accept. In any case, these models are not based
on
any ideas about ancestry of the organisms studied, so the taxonomy produced
is not systematic. In a phenetic system, one might measure flower color
as one
characteristic. It is useful for separating flowering plants into groups
when using phenetic methods.
However, this may not be enough to qualify flower color as a useful systematic
characteristic.
It is useful in systematics only if those flowers with the same color
are similar because they are descended from an ancestor with that color flower.
Thus,
flower
color is determined by ancestry (by the way, flower color is sometimes useful
and sometimes not to systemetists).
Cladistics is a particular systematic scheme based on an explicit
theory of evolution. In cladistics., organisms inherit their genes
through meiosis
or mitosis (so it applies to sexual and asexual organisms). This is the
basis for the focus on ancestry as linking organisms into groups. As change
occurs,
a group of organisms splits into two different branches (= clades,
hence the odd name for this method). Thus, a group grows like a tree,
with the ancestral
group being the trunk. As you go from the base of the tree upward, you
are traveling through time towards the present. The trunk splits (= bifurcates
because the
splits are always into two new branches) and the original group is now replaced
with two new groups, each of which may split, so the tree grows more
and more
branches as time passes. The tips of the last splits (the tiniest twigs
on the tree) represent the most recent clades, the only clades still in existence. The tree metaphor is a very good one. In fact, the result of a
cladistic analysis is a tree-like diagram called a dendrogram
(the root term "dendro" comes from the Greek dendron for
tree).
Cladistics is a set of methods
to uncover the tree, which is all in the past (by definition), using characteristics
measured from the organisms alive today. Since the methods are standardized,
the same data will produce the same groupings. There are a lot of different
methodologies, but the problem of multiple methods is not as difficult for
cladists as it is for pheneticists. This is because systematics is a
natural system,
so the different methods are all uncovering the same natural groups. Thus,
the assumption is that they should all produce the same results if the data
and
methods are free of error. To construct a cladogram, one must measure
characteristics that indicate common ancestry. Whale flippers, primate
hands, and frog paws
all have five fingers. The development of these organs supports the hypothesis
that they are similar because the ancestor of these organisms had five fingers.
You may recognize this similarity-due-to-descent as homology. Lets
go back to flower color. Flowers in cool tropical uplands are more often
red than
expected.
Is this because the plants in this habitat are all related? In
this case, probably not. Lots of very different plants occur in these
areas. However, cool
areas
like these have fewer flying insects and many pollinators are not insects but
vertebrates. Reds are apparently easier for vertebrates to see so the
increased
incidence of red flowers is probably related to the utility of having a flower
that is easily seen. In this habitat, red flower color does not necessarily
indicate
common ancestry.
Deciding which characteristics
contain information about ancestry can be tricky. This, and the large
number of calculations needed to analyze data, limited cladistic analyses
for many
years. However, two technological advances have turned it into a preferred
means of making natural classifications. First, computers have made
it possible to
do the many trillions of calculations often needed to perform an analysis.
Second, DNA sequencing technology has given researchers the means
to measure characteristics
(gene sequences) that are the very stuff of common descent. The details
can get a bit tricky but, in general, the assumption that two individuals have
the
same gene sequence because they share a common ancestor is a reasonable one.
How,
you might ask, does all of this affect you in this class?
In articles, book and websites devoted to plant classification, you will
see that it either uses a standard taxonomic or a cladistic classification. You
have to recognize which is being used and to appreciate the reasons why the
two schemes are not equivalents. If you go to the USDA site to classify
your
plants, you will get a standard taxonomic approach, where there are species,
genera, families, etc. "The Tree of Life" site (notice the use of
the metaphorical tree again) is a second place to get an overall classification
of plants that uses a cladistic scheme. They are not the same. Why?
The USDA site is based on a taxonomic scheme that tries to fit plants
into a preset
hierarchy.
Those who have constructed this hierarchy (many many researchers working
over many many years) have tried to use characteristics that reflect common
ancestry
(so this is an attempt at a natural system) but made groups according to a
pre-ordained taxonomic structure. This approach is called evolutionary
systematics (a bit
of a redundancy). Thus, all plants in the family Fabaceae (pea family)
are believed to share a common ancestor, i. e. to be more related to one another
than to
other plants. The "Tree of Life" site presents a strictly cladistic
analysis, in which the branching pattern arises from the data, not from a
preset
taxonomic hierarchy. A cladogram is not the same as the standard taxonomic
hierarchy and can be very different from it. Just try it for the plants.
The USDA site
presents the plants like an outline, in which each lower taxonomic level is
indented from the next higher level. Go
there and see at http://plants.usda.gov/cgi_bin/topics.cgi?earl=classification.html.
Select "Class" as the rank and you will get a hierarchical
classification of all plants to the level of the family. I have reproduced
the results below.
Kingdom Plantae
-- Plants
- Division Anthocerotophyta
-- Hornworts
- Division Bryophyta
-- Mosses
- Subdivision Musci
- Class Andreaeopsida
-- Granite mosses
- Class Bryopsida
-- True mosses
- Class Sphagnopsida
-- Peat mossed
- Division Hepatophyta
-- Liverworts
- Subkingdom Tracheobionta
-- Vascular plants
- Division Equisetophyta
-- Horsetails
- Division Lycopodiophyta
-- Lycopods
- Division Psilophyta
-- Whisk-ferns
- Division Pteridophyta
-- Ferns
- Superdivision Spermatophyta
-- Seed plants
- Division Coniferophyta
-- Conifers
- Division Cycadophyta
-- Cycads
- Division Ginkgophyta
-- Ginkgo
- Division Gnetophyta
-- Mormon tea and other gnetophytes
- Division Magnoliophyta
-- Flowering plants
- Class Liliopsida
-- Monocotyledons
- Class Magnoliopsida
-- Dicotyledons
If you go the the Tree
of Life web page on embryophytes, the nearest you can to
the level presented in the USDA above, you get the cladogram presented below.
Notice that there are some similar names with different suffixes, like
Filicopsida below and above or Lycopodiopsida above and Lycopsida below. Notice
that both
are classes above, but what are they below? "Lycopsid" appears
at a different level of the dendrogram than does "Filicopsida." What
does "class" mean in the dendrogram below? It turns out that
there is no equivalent to class (or family, or order) in the Tree of Life's
cladistic
approach. So, even though "Filicopsida" includes the same plants
in both schemes, it fits into the overall scheme very differently in each
approach
to taxonomy.
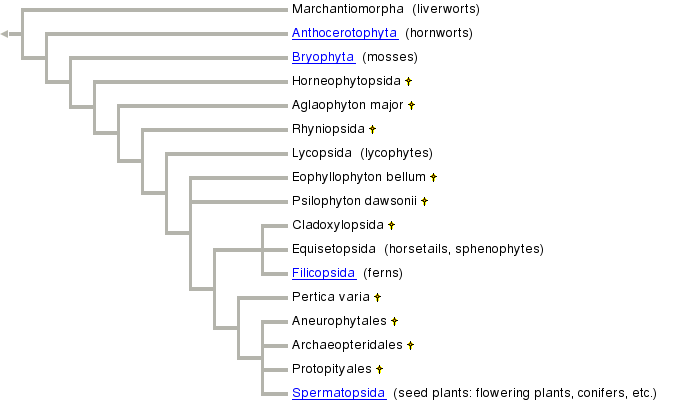
The conclusion to draw from this is that
there are different taxonomic methods to serve different purposes. The
standard hierarchy is preferred as an organizational scheme - a good filing
system. Using
it, one can quickly find plants with particular characteristics. The
standardization of the hierarchy (Kingdom, Division, etc.) is familiar and
easy to visualize.
However, it is not a truly systematic approach. Attempts to make
it more natural fail when the actual groups can't be easily fitted into the
hierarchical
scheme
without making loads of new levels (superfamilies, subclasses, sections, groups,
etc.) which makes the scheme too cumbersome. Cladistics are the best
method
we have of presenting the evolutionary history of an organism because of the
flexibility of the bifurcating tree. However, the trees are not easy
to memorize
and it may be difficult to find any phenotypic (morphological, anatomical or
chemical) characteristics that separate and uniquely define a group that
is
formed by analyzing the genotype (the gene sequences). In addition, there
are evolutionary events that are difficult to represent with a bifurcating
tree
(new species arising from hybridization is one problem). No scheme is
perfect at this time and a perfect scheme is unlikely in the future.
A note on the taxonomic levels used in
botanical classification. Most should be familiar to you from animal
taxonomy: species, genus, family, order, and class. The only difference
is the use of
"division" by botanists
rather than "phylum"
for the level below that of kingdom. There is a reason for this change.
A phylum represents a basic "body plan" in the animal kingdom.
All rotifers have key characteristics that form the rotifer body plan,
as all chordates
must
have the basic chordate body plan. However, related plants do not seem
to have a basic body plan. The pea family contains herbaceous members,
like garden
beans
and peas, members that are bushes (Gorse is an example of a bush pea), and
members that are trees, like the honey locust trees that bloom so profusely
in Tennessee. So,
what's the body plan here? Hard to say, so the
idea that plants can be separated into basic body plans never took hold and
the use of the turn "phylum" would be inappropriate. Instead,
classic plant taxonomy relied on the anatomy of the reproductive structures
(that's
how locust trees and beans got into the same groups - just look at the pods
they each produce).
There is an excellent free resource for
learning more about cladistics. "The Compleat Cladist" (the title
is a take-off of Izaak Walton's 17th century meditation on fishing, "The
Compleat Angler" - hence the odd spelling) by E. O. Wiley, D. Siegel-Causey,
D. R. Brooks, and V. A. Funk (1991, Special Publication No. 19, The University
of Kansas Museum of Natural History, Lawrence, KS) is both a primer on how
to construct trees and a good introduction to the theory behind cladism.
Since
there is to be a second edition, the authors have been distributing the entire
book as a pdf file. It can be obtained at or by emailing me and
requesting I
send a copy of my copy. As with any pdf file, you need a reader application
to view it. The easiest to get is Acrobat Reader from http://www.adobe.com
if you computer doesn't already have it. It is freeware.
Last updated July 9, 2013


